Museomics to study insect population decline
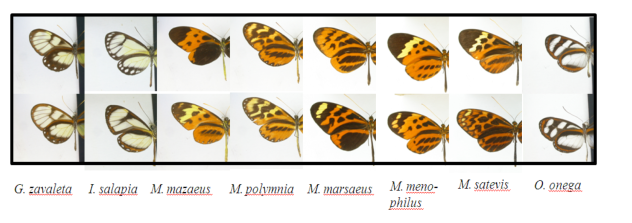
Population genomics of mimetic butterfly from hybrid zones
During my previous post-doc at the Inria of Rennes in collaboration with the National Museum of Natural History of Paris, I studied different mimetic butterfly species (Nymphalidae, Danainae, Ithominii) showing diverging populations and hybridization across a suture zone in the Andean region. Using population genomics I tried to shape the evolutionary history of these populations.
Adaptation genomics of the Cotesia parasitoid wasps
The Cotesia parasitic wasps comprise several species that parasitize Lepidoptera caterpillars. Larvae grow inside their host and to overcome host defenses these wasps developed an original strategy: the domestication of viruses, named bracoviruses (BVs). These bracoviruses have been stably integrated in the genome of the wasps and play a crucial role in the wasp life cycle as they produce bracovirus particles, which are injected into parasitized lepidopteran hosts during wasp oviposition and are necessary for successful development of wasp larvae within the host (Gauthier et al. 2017 Parasitology for review).

During my thesis, I have been fully involved in the ANR ABC-Papogen project whose aim was to understand the ecological adaptation of the parasitoid wasp Cotesia sesamiae at different evolutionary levels.
A first axis aimed to obtain a high quality reference genome for the wasp Cotesia congregata, to undertake a comparative genomics approach with 6 other wasp species. This part required the creation of a consortium I animated, to annotate different functions related to the parasitic success and the ability to find, select and attack hosts. The genomic data allowed to study the evolution of multigenic families , gene expansions, selection pressures etc. but also their genomic architecture (Gauthier et al. in prep).
The second axis was focused on the population genomics of the wasp using cutting edge sequencing techniques (such as targeted resequencing and RAD sequencing) based on a large field sampling from Africa. These approaches highlights the determinant influence of host adaptation in the population evolutionary trajectories (Kaiser et al. 2015 Evolutionary Applications ; Branca et al. 2018 biorxiv) including the emergence of a new species Cotesia typhae (Kaiser et al. 2017 Zookeys). Moreover, in this context of strong interaction with the host range, the bracovirus seems to play a crucial role (Gauthier et al. 2018 Molecular Ecology).
